Chapter Contents
Previous
Next
|
Chapter Contents |
Previous |
Next |
| The CALIS Procedure |
McDonald (1980) uses the same data set to compute a factor analysis with ordinally constrained factor loadings. The results of the linearly constrained factor analysis show that the loadings of the two factors are ordered as 2, 1, 3, 4, 6, 5. McDonald (1980) then tests the hypothesis that the factor loadings are all nonnegative and can be ordered in the following manner:
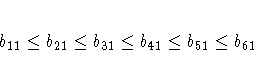
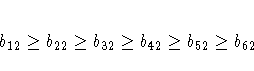
proc calis data=Kinzer method=max outram=ram tech=nr nobs=326;
Title2 "Ordinally Related Factor Analysis, (Mcdonald,1980)";
Title3 "Identification Problem";
Cosan F(8,Gen) * I(8,Ide);
MATRIX F
[,1] = x1-x6,
[,2] = x7-x12,
[1,3] = x13-x18;
PARAMETERS t1-t10=1.;
x2 = x1 + t1 * t1;
x3 = x2 + t2 * t2;
x4 = x3 + t3 * t3;
x5 = x4 + t4 * t4;
x6 = x5 + t5 * t5;
x11 = x12 + t6 * t6;
x10 = x11 + t7 * t7;
x9 = x10 + t8 * t8;
x8 = x9 + t9 * t9;
x7 = x8 + t10 * t10;
Bounds x13-x18 >= 0.;
Vnames F Fact1 Fact2 Uvar1-Uvar6;
run;
You can specify the same model with the LINCON statement:
proc calis data=Kinzer method=max tech=lm edf=325;
Title3 "Identified Problem 2";
cosan f(8,gen)*I(8,ide);
matrix F
[,1] = x1-x6,
[,2] = x7-x12,
[1,3] = x13-x18;
lincon x1 <= x2,
x2 <= x3,
x3 <= x4,
x4 <= x5,
x5 <= x6,
x7 >= x8,
x8 >= x9,
x9 >= x10,
x10 >= x11,
x11 >= x12;
Bounds x13-x18 >= 0.;
Vnames F Fact1 Fact2 Uvar1-Uvar6;
run;
To have an identified model, the loading, b11 (x1), is fixed at 0. The information in the OUTRAM= data set (the data set ram), produced by the unidentified model, can be used to specify the identified model. However, because x1 is now a fixed constant in the identified model, it should not have a parameter name in the new analysis. Thus, the data set ram is modified as follows:
data ram2(type=ram); set ram;
if _name_ = 'x1' then do;
_name_ = ' '; _estim_ = 0.;
end;
run;
The data set ram2 is now an OUTRAM= data set in which x1 is no longer a parameter. PROC CALIS reads the information (that is, the set of parameters and the model specification) in the data set ram2 for the identified model. As displayed in the following code, you can use the PARMS statement to specify the desired ordinal relationships between the parameters.
proc calis data=Kinzer method=max inram=ram2 tech=nr nobs=326;
title2 "Ordinally Related Factor Analysis, (Mcdonald,1980)";
title3 "Identified Model with X1=0";
parms t1-t10= 10 * 1.;
x2 = + t1 * t1;
x3 = x2 + t2 * t2;
x4 = x3 + t3 * t3;
x5 = x4 + t4 * t4;
x6 = x5 + t5 * t5;
x11 = x12 + t6 * t6;
x10 = x11 + t7 * t7;
x9 = x10 + t8 * t8;
x8 = x9 + t9 * t9;
x7 = x8 + t10 * t10;
bounds x13-x18 >= 0.;
run;
Selected output for the identified model is displayed in Output 19.5.1.
Output 19.5.1: Factor Analysis with Ordinal Constraints
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The singular Hessian matrix of the unidentified problem slows down the convergence rate of the Levenberg-Marquardt algorithm considerably. Compared to the unidentified problem with 30 iterations, the identified problem needs only 20 iterations. Note that the number of iterations may depend on the precision of the processor.
The same model can also be specified using the LINCON statement for linear constraints:
proc calis data=Kinzer method=max tech=lm edf=325;
Title3 "Identified Model 2";
cosan f(8,gen)*I(8,ide);
matrix f
[,1] = 0. x2-x6,
[,2] = x7-x12,
[1,3] = x13-x18;
lincon x2 <= x3,
x3 <= x4,
x4 <= x5,
x5 <= x6,
x7 >= x8,
x8 >= x9,
x9 >= x10,
x10 >= x11,
x11 >= x12;
bounds x2 x13-x18 >= 0.;
run;
|
Chapter Contents |
Previous |
Next |
Top |
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.