PROC MDS Statement
- PROC MDS < options > ;
By default, the only result produced by the MDS procedure is the
iteration history. Hence, you should always specify one or
more options for output data sets (OUT=, OUTFIT=, and OUTRES=)
or displayed output (such as PFINAL). PROC MDS does not produce
any plots; to produce plots, use the output data sets with PROC
PLOT or PROC GPLOT.
The types of estimates written to the OUT= data set
are determined by the OCONFIG, OCOEF, OTRANS, and OCRIT options.
If you do not specify any of these four options, the estimates
of all the parameters of the PROC MDS model and the value of the
badness-of-fit criterion appear in the OUT= data set. If you
specify one or more of these options, only the information
requested by the specified options appear in the OUT= data set.
Also, the OITER option causes these statistics to be written to the
OUT= data set after initialization and on each iteration, as well as
after the iterations have terminated.
Displayed output is controlled by the interaction of the
PCONFIG, PCOEF, PTRANS, PFIT, and PFITROW options with
the PININ, PINIT, PITER, and PFINAL options.
The PCONFIG, PCOEF, PTRANS, PFIT, and PFITROW options specify which
estimates and fit statistics are to be displayed. The PININ, PINIT,
PITER, and PFINAL options specify when the estimates and fit
statistics are to be displayed. If you specify at least one of
the PCONFIG, PCOEF, PTRANS, PFIT and PFITROW options but none
of the PININ, PINIT, PITER, and PFINAL options, the final
results (PFINAL) are displayed. If you specify at least one of
the PININ, PINIT, PITER, and PFINAL options but none of the
PCONFIG, PCOEF, PTRANS, PFIT and PFITROW options, all estimates
(PCONFIG, PCOEF, PTRANS) and the fit statistics for each matrix
and for the entire sample (PFIT) are displayed. If you do not
specify any of these nine options, no estimates or fit statistics
are displayed (except the badness-of-fit criterion in the iteration
history).
- ALTERNATE | ALT=NONE | NO | N
- ALTERNATE | ALT=MATRIX | MAT | M | SUBJECT | SUB | S
- ALTERNATE | ALT=ROW | R <=n>
-
determines what form of alternating-least-squares algorithm is used.
The default depends on the amount of memory available.
The following ALTERNATE= options are listed in order of decreasing memory
requirements:
- ALT=NONE
- causes all parameters to be adjusted simultaneously
on each iteration. This option is usually best for a small number
of subjects and objects.
- ALT=MATRIX
- adjusts all the parameters for the first subject,
then all the parameters for the second subject, and so on, and
finally adjusts all parameters that do not correspond to a subject,
such as coordinates and unconditional transformations. This option
usually works best for a large number of subjects with a small number
of objects.
- ALT=ROW
- treats subject parameters the same way as the
ALTERNATE=MATRIX option but also includes separate stages for
unconditional parameters and for subsets of the objects.
The ALT=ROW option usually works best for a large number of objects.
Specifying ALT=ROW=n divides the objects into subsets of n
objects
each, except possibly for one subset when n does not divide the
number of objects evenly. If you omit =n, the number of objects
in the subsets is determined from the amount of memory available.
The smaller the value of n, the less memory is required.
When you specify the LEVEL=ORDINAL option,
the monotone transformation is always
computed in a separate stage and is listed as a separate
iteration in the iteration history. In this case, estimation is done
by iteratively reweighted least squares. The weights are recomputed
according to the FORMULA= option on each monotone iteration; hence,
it is possible for the badness-of-fit criterion to increase after
a monotone iteration.
- COEF=IDENTITY | IDEN | I
- COEF=DIAGONAL | DIAG | D
-
specifies the type of matrix for the dimension coefficients.
- COEF=IDENTITY
- is the default, which yields Euclidean distances.
- COEF=DIAGONAL
- produces weighted Euclidean distances, in which
each subject is allowed differential weights for the dimensions.
The dimension coefficients that PROC MDS outputs are related to
the square roots of what are called subject weights in PROC ALSCAL;
the normalization in PROC MDS also differs from that in PROC ALSCAL.
The weighted Euclidean model is related to the INDSCAL model
(Carroll and Chang 1970).
- CONDITION | COND=UN | U
- CONDITION | COND=MATRIX | MAT | M | SUBJECT | SUB | S
- CONDITION | COND=ROW | R
-
specifies the conditionality of the data (Young 1987, pp. 60-63).
The default is CONDITION=MATRIX.
The data are divided into disjoint subsets called partitions.
Within each partition, a separate transformation is applied, as
specified by the LEVEL= option.
- COND=UN
- puts all the data into a single partition.
- COND=MATRIX
- makes each data matrix a partition.
- COND=ROW
- makes each row of each data matrix a partition.
The CONDITION= option also determines the default value for the
SHAPE= option. If you specify the CONDITION=ROW option and omit the
SHAPE= option, each data
matrix is stored as a square and possibly asymmetric matrix.
If you specify the CONDITION=UN or CONDITION=MATRIX option and omit the
SHAPE= option, only one triangle
is stored. See the
SHAPE= option for details.
- CONVERGE | CONV=p
-
sets both the gradient convergence criterion and the monotone
convergence criterion to p, where
 . The default is CONVERGE=.01; smaller values may greatly increase
the number of iterations required. Values less than .0001 may be
impossible to satisfy because of the limits of machine precision.
See the GCONVERGE=
and MCONVERGE= options.
. The default is CONVERGE=.01; smaller values may greatly increase
the number of iterations required. Values less than .0001 may be
impossible to satisfy because of the limits of machine precision.
See the GCONVERGE=
and MCONVERGE= options.
- CUTOFF=n
-
causes data less than n to be replaced by missing values.
The default is CUTOFF=0.
- DATA=SAS-data-set
-
specifies the SAS data set containing one or more square matrices
to be analyzed. In typical psychometric data, each matrix contains
judgments from one subject, so there is a one-to-one correspondence
between data matrices and subjects.
The data matrices contain similarity or dissimilarity measurements
to be modeled and, optionally, weights for these data. The data are
generally assumed to be dissimilarities unless you use the SIMILAR
option. However, if there are nonmissing diagonal values and these
values are predominantly larger than the off-diagonal values, the
data are assumed to be similarities and are treated as if the
SIMILAR option is specified. The diagonal elements are not
otherwise used in fitting the model.
Each matrix must have exactly the same number of observations as
the number of variables specified by the VAR statement or
determined by defaults. This number is the number of objects or
stimuli.
The first observation and variable are assumed to contain data
for the first object, the second observation and variable are assumed to
contain data for the second object, and so on.
When there are two or more matrices, the observations in each matrix
must correspond to the same objects in the same order as in the
first matrix.
The matrices can be symmetric or asymmetric, as specified by the
SHAPE= option.
- DECIMALS | DEC=n
-
specifies how many decimal places to use when displaying the parameter
estimates and fit statistics. The default is DECIMALS=2, which is
generally reasonable except in conjunction with the LEVEL=ABSOLUTE
option and very large
or very small data.
- DIMENSION | DIMENS | DIM=n < TO m < BY=i >>
-
specifies the number of dimensions to use in the MDS model,
where
 number of objects.
The parameter i can be either positive or negative but
not zero. If you
specify different values for n and m, a
separate model is fitted for each requested dimension.
If you specify only
DIMENSION=n, then only n dimensions are fitted.
The default is DIMENSION=2 if there are three or more objects;
otherwise, DIMENSION=1 is the only valid specification. The
analyses for each number of dimensions are done independently.
For information on choosing the dimensionality, refer to Kruskal
and Wish (1978, pp. 48-60).
number of objects.
The parameter i can be either positive or negative but
not zero. If you
specify different values for n and m, a
separate model is fitted for each requested dimension.
If you specify only
DIMENSION=n, then only n dimensions are fitted.
The default is DIMENSION=2 if there are three or more objects;
otherwise, DIMENSION=1 is the only valid specification. The
analyses for each number of dimensions are done independently.
For information on choosing the dimensionality, refer to Kruskal
and Wish (1978, pp. 48-60).
- EPSILON | EPS=n
-
specifies a number n, 0 < n < 1, that determines
the amount added to squared distances computed from the model
to avoid numerical problems such as division by 0. This amount
is computed as
 equal to n times
the mean squared distance in the initial configuration. The
distance in the MDS model is thus computed as
equal to n times
the mean squared distance in the initial configuration. The
distance in the MDS model is thus computed as
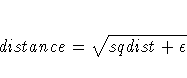
where sqdist is the squared Euclidean distance or the
weighted squared Euclidean distance.
The default is EPSILON=1E-12, which is small enough to have no
practical effect on the estimates unless the FIT= value is
nonpositive and there are dissimilarities that are very close to 0.
Hence, when the FIT= value is nonpositive, dissimilarities
less than n times 100 times the maximum dissimilarity
are disallowed.
- FIT=DISTANCE | DIS | D
- FIT=SQUARED | SQU | S
- FIT=LOG | L
- FIT=n
-
specifies a predetermined (not estimated) transformation to
apply to both sides of the MDS model before the error term
is added.
The default is FIT=DISTANCE or, equivalently, FIT=1, which fits
data to distances.
The option FIT=SQUARED or FIT=2 fits squared data to squared distances.
This gives greater importance to large data and distances and
lesser importance to small data and distances in fitting the
model.
The FIT=LOG or FIT=0 option fits log data to log distances.
This gives lesser importance to large data and distances and
greater importance to small data and distances in fitting the model.
In general, the FIT=n option fits nth-power data to
nth-power distances. Values of n that are
large in absolute value can cause floating-point overflows.
If the FIT= value is 0 or negative, the data must be strictly
positive (see the EPSILON= option). Negative data may produce
strange results with any value other than FIT=1.
- FORMULA | FOR=0 | OLS | O
- FORMULA | FOR=1 | USS | U
- FORMULA | FOR=2 | CSS | C
-
determines how the badness-of-fit criterion is standardized
in correspondence
with stress formulas 1 and 2 (Kruskal and Wish 1978, pp. 24-26).
The default is FORMULA=1 unless you specify FIT=LOG, in which
case the default is FORMULA=2. Data partitions are
defined by the CONDITION= option.
- FORMULA=0
- fits a regression model by ordinary least squares (Null and Sarle 1982)
without standardizing the partitions; this option cannot be used with
the LEVEL=ORDINAL option. The badness-of-fit criterion is the square root
of the error sum of squares.
- FORMULA=1
- standardizes each partition by the uncorrected sum of
squares of the (possibly transformed) data; this option should not be
used with the FIT=LOG option. With the FIT=DISTANCE and
LEVEL=ORDINAL options, this
is equivalent to Kruskal's stress formula 1 or an obvious
generalization thereof. With the FIT=SQUARED and LEVEL=ORDINAL options,
this is equivalent to Young's s-stress formula 1 or an obvious
generalization thereof. The badness-of-fit criterion is analogous
to
 , where R is a multiple correlation
about the origin.
, where R is a multiple correlation
about the origin.
- FORMULA=2
- standardizes each partition by the corrected sum of
squares of the (possibly transformed) data; this option is the recommended
method for unfolding. With the FIT=DISTANCE and LEVEL=ORDINAL options, this
is equivalent to Kruskal's stress formula 2 or an obvious
generalization thereof. With the FIT=SQUARED and LEVEL=ORDINAL options,
this is equivalent to Young's s-stress formula 2 or an obvious
generalization thereof. The badness-of-fit criterion is analogous
to
 , where R is a multiple correlation
computed with a denominator corrected for the mean.
, where R is a multiple correlation
computed with a denominator corrected for the mean.
- GCONVERGE | GCONV=p
-
sets the gradient convergence criterion to p, where
 . The default is GCONVERGE=0.01; smaller
values may greatly increase the number of iterations required.
Values less than 0.0001 may be impossible to satisfy because
of the limits of machine precision.
. The default is GCONVERGE=0.01; smaller
values may greatly increase the number of iterations required.
Values less than 0.0001 may be impossible to satisfy because
of the limits of machine precision.
The gradient convergence measure is the multiple correlation
of the Jacobian matrix with the residual vector, uncorrected
for the mean.
See the CONVERGE=
and MCONVERGE= options.
- INAV=DATA | D
- INAV=SSCP | S
-
affects the computation of initial coordinates. The default is INAV=DATA.
- INAV=DATA
- computes a weighted average of the data matrices. Its value is
estimated only if an element is missing from every data matrix.
The weighted average of the data matrices
with missing values filled in is then converted to a scalar products
matrix (or what would be a scalar products matrix if the fit were
perfect), from which the initial coordinates are computed.
- INAV=SSCP
- estimates missing values in each data matrix and
converts each data matrix to a scalar products matrix. The initial
coordinates are computed from the unweighted average of the
scalar products matrices.
- INITIAL | IN=SAS-data-set
-
specifies a SAS data set containing initial values for some or
all of the parameters of the MDS model.
If the INITIAL= option is omitted, the initial values are
computed from the data.
- LEVEL=ABSOLUTE | ABS | A
- LEVEL=RATIO | RAT | R
- LEVEL=INTERVAL | INT | I
- LEVEL=LOGINTERVAL | LOG | L
- LEVEL=ORDINAL | ORD | O
-
specifies the measurement level of the data and hence the
type of estimated (optimal) transformations applied to the
data or distances
(Young 1987, pp. 57-60; Krantz et. al. 1971, pp. 9-12)
within each partition as specified by the CONDITION= option.
The default is LEVEL=ORDINAL.
- LEVEL=ABSOLUTE
- allows no optimal transformations. Hence, the
distinction between regression and measurement models is
irrelevant.
- LEVEL=RATIO
- fits a regression model in which the distances are
multiplied by a slope parameter in each partition (a linear
transformation). In this case, the regression model is equivalent
to the measurement model with the slope parameter reciprocated.
- LEVEL=INTERVAL
- fits a regression model in which the distances are
multiplied by a slope parameter and added to an intercept
parameter in each partition (an affine transformation). In this
case, the regression and measurement models differ if there is
more than one partition.
- LEVEL=LOGINTERVAL
- fits a regression model in which the distances are
raised to a power and multiplied by a slope parameter in each
partition (a power transformation).
- LEVEL=ORDINAL
- fits a measurement model in which a least-squares
monotone increasing transformation is applied to the data in each
partition. At the ordinal measurement level, the regression and
measurement models differ.
- MAXITER | ITER=n
-
specifies the maximum number of iterations, where
 . The default is MAXITER=100.
. The default is MAXITER=100.
- MCONVERGE | MCONV=p
-
sets the monotone convergence criterion to p,
where
 , for use with the LEVEL=ORDINAL option.
The default is MCONVERGE=0.01; if you want greater precision,
MCONVERGE=0.001 is usually reasonable, but smaller values may
greatly increase the number of iterations required.
, for use with the LEVEL=ORDINAL option.
The default is MCONVERGE=0.01; if you want greater precision,
MCONVERGE=0.001 is usually reasonable, but smaller values may
greatly increase the number of iterations required.
The monotone convergence criterion is the Euclidean norm of the
change in the optimally scaled data divided by the Euclidean
norm of the optimally scaled data, averaged across partitions
defined by the CONDITION= option.
See the CONVERGE=
and GCONVERGE= options.
- MINCRIT | CRITMIN=n
-
causes iteration to terminate when the badness-of-fit criterion
is less than or equal to n, where
 . The default is MINCRIT=1E-6.
. The default is MINCRIT=1E-6.
- NEGATIVE
-
allows slopes or powers to be negative with the LEVEL=RATIO,
INTERVAL, or LOGINTERVAL option.
- NONORM
-
suppresses normalization of the initial and final estimates.
- NOPHIST | NOPRINT | NOP
-
suppresses the output of the iteration history.
- NOULB
-
causes missing data to be estimated during initialization by the
average nonmissing value, where the average is computed
according to the FIT= option. Otherwise, missing data are
estimated by interpolating between the Rabinowitz (1976) upper and
lower bounds.
- OCOEF
-
writes the dimension coefficients to the OUT= data set.
See the OUT= option for interactions with other options.
- OCONFIG
-
writes the coordinates of the objects to the OUT= data set.
See the OUT= option for interactions with other options.
- OCRIT
-
writes the badness-of-fit criterion to the OUT= data set.
See the OUT= option for interactions with other options.
- OITER | OUTITER
-
writes current values to the output data sets after initialization
and on every iteration. Otherwise, only the final values are
written to any output data sets.
See the OUT=, OUTFIT=, and OUTRES= options.
- OTRANS
-
writes the transformation parameter estimates to the OUT= data set
if any such estimates are computed. There are no transformation
parameters with the LEVEL=ORDINAL option. See the OUT= option
for interactions with other options.
- OUT=SAS-data-set
-
creates a SAS data set containing, by default, the estimates
of all the parameters of the PROC MDS model and the value of the
badness-of-fit criterion. However, if you specify one or more
of the OCONFIG, OCOEF, OTRANS, and OCRIT options, only the
information requested by the specified options appears in the
OUT= data set. See also the OITER option.
- OUTFIT=SAS-data-set
-
creates a SAS data set containing goodness-of-fit and badness-of-fit
measures for each partition as well as for the entire data set.
See also the OITER option.
- OUTRES=SAS-data-set
-
creates a SAS data set containing one observation for each
nonmissing datum from the DATA= data set. Each observation
contains the original datum, the estimated distance computed
from the MDS model, transformed data and distances, and the
residual.
See also the OITER option.
- OVER=n
-
specifies the maximum overrelaxation factor,
where
 . Values between 1 and 2 are
generally reasonable. The default is OVER=2 with
the LEVEL=ORDINAL, ALTERNATE=MATRIX, or
ALTERNATE=ROW option; otherwise, the default is OVER=1.
Use this option only if you have convergence problems.
. Values between 1 and 2 are
generally reasonable. The default is OVER=2 with
the LEVEL=ORDINAL, ALTERNATE=MATRIX, or
ALTERNATE=ROW option; otherwise, the default is OVER=1.
Use this option only if you have convergence problems.
- PCOEF
-
produces the estimated dimension coefficients.
- PCONFIG
-
produces the estimated coordinates of the objects in the configuration.
- PDATA
-
displays each data matrix.
- PFINAL
-
displays final estimates.
- PFIT
-
displays the badness-of-fit criterion and various types of
correlations between the data and fitted values for each
data matrix, as well as for the entire sample.
- PFITROW
-
displays the badness-of-fit criterion and various types of
correlations between the data and fitted values for each
row as well as for each data matrix and for the entire
sample. This option works only with the CONDITION=ROW option.
- PINAVDATA
-
displays the sum of the weights and the weighted average of the data
matrices computed during initialization with the INAV=DATA option.
- PINEIGVAL
-
displays the eigenvalues computed during initialization.
- PINEIGVEC
-
displays the eigenvectors computed during initialization.
- PININ
-
displays values read from the INITIAL= data set. Since these
values may be incomplete, the PFIT and PFITROW options do not apply.
- PINIT
-
displays initial values.
- PITER
-
displays estimates on each iteration.
- PTRANS
-
displays the estimated transformation parameters if any are computed.
There are no transformation parameters with the LEVEL=ORDINAL option.
- RANDOM<=seed>
-
causes initial coordinate values to be pseudorandom numbers.
In one dimension, the pseudorandom numbers are uniformly
distributed on an interval.
In two or more dimensions, the pseudorandom numbers are uniformly
distributed on the circumference of a circle or the surface of a
(hyper)sphere.
- RIDGE=n
-
specifies the initial ridge value, where
 . The default is RIDGE=1E-4.
. The default is RIDGE=1E-4.
If you get a floating-point overflow in the first few iterations,
specify a larger value such as RIDGE=0.01 or RIDGE=1 or RIDGE=100.
If you know that the initial estimates are very good, using RIDGE=0
may speed convergence.
- SHAPE=TRIANGULAR | TRIANGLE | TRI | T
- SHAPE=SQUARE | SQU | S
-
determines whether the entire data matrix for each subject
or only one triangle of the matrix is
stored and analyzed.
If you specify the CONDITION=ROW option, the default is SHAPE=SQUARE.
Otherwise, the default is SHAPE=TRIANGLE.
- SHAPE=SQUARE
- causes the entire matrix to be stored and analyzed.
The matrix can be asymmetric.
- SHAPE=TRIANGLE
- causes only one triangle to be stored. However,
PROC MDS reads both upper and lower triangles to look for
nonmissing values and to symmetrize the data if needed. If corresponding
elements in the upper and lower triangles both contain nonmissing
values, only the average of the two values is stored and analyzed
(Kruskal and Wish 1978, p. 74).
Also, if an OUTRES= data set is requested, only the average of the
two corresponding elements is output.
- SIMILAR | SIM<=max>
-
causes the data to be treated as similarity measurements rather
than dissimilarities. If =max is not specified, each datum is
converted to a dissimilarity by subtracting it from the maximum
value in the data set or BY group. If =max is specified,
each datum is subtracted from the maximum of max and the
data. The diagonal data are included in computing these maxima.
By default, the data are assumed to be dissimilarities unless
there are nonmissing diagonal values and these values are
predominantly larger than the off-diagonal values. In this case,
the data are assumed to be similarities and are treated as if the
SIMILAR option is specified.
- SINGULAR=p
-
specifies the singularity criterion p,
 . The default is SINGULAR=1E-8.
. The default is SINGULAR=1E-8.
- UNTIE
-
allows tied data to be assigned different optimally scaled values
with the LEVEL=ORDINAL option. Otherwise, tied data are assigned
equal optimally scaled values. The UNTIE option has no effect with
values of the LEVEL= option other than LEVEL=ORDINAL.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.