Computational Formulas
Product-Limit Method
Let t1 < t2 < ... < tk represent the distinct event times.
For each i = 1, ... ,k, let ni be the number of surviving
units, the size of the risk set, just prior to ti.
Let di be the number of units that
fail at ti, and let si=ni-di.
The product-limit estimate of the SDF
at ti is the cumulative product

Notice that the estimator is defined to be right continuous; that
is, the events at ti are included in the estimate of S(ti).
The corresponding estimate of the standard error is computed
using Greenwood's formula (Kalbfleish and Prentice 1980) as
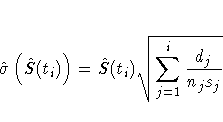
The first sample quartile of the
survival time distribution is given by

Confidence intervals for the quartiles are based on the sign
test (Brookmeyer and Crowley 1982). The  confidence
interval for the first quartile is given by
confidence
interval for the first quartile is given by

where 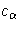 is the upper
is the upper  percentile of a central chi-squared
distribution with 1 degree of freedom.
The second and third sample quartiles and the corresponding
confidence intervals are calculated by replacing the 0.25 in the last two
equations by 0.50 and
0.75, respectively.
percentile of a central chi-squared
distribution with 1 degree of freedom.
The second and third sample quartiles and the corresponding
confidence intervals are calculated by replacing the 0.25 in the last two
equations by 0.50 and
0.75, respectively.
The estimated mean survival time is

where t0 is defined to be zero.
If the last observation is censored,
this sum underestimates the mean.
The standard error of  is estimated as
is estimated as

where

Life Table Method
The life table estimates are computed by counting the
numbers of censored and uncensored observations that
fall into each of the time intervals [ti-1,ti),
i = 1,2, ... ,k+1, where t0=0 and  .Let ni be the number of units entering the
interval [ti-1,ti), and let di be the
number of events occurring in the interval.
Let bi=ti-ti-1, and let ni' = ni - wi/2,
where wi is the number of units censored in the interval.
The effective sample size of the interval [ti-1,ti) is
denoted by ni'.
Let tmi denote the midpoint of [ti-1,ti).
.Let ni be the number of units entering the
interval [ti-1,ti), and let di be the
number of events occurring in the interval.
Let bi=ti-ti-1, and let ni' = ni - wi/2,
where wi is the number of units censored in the interval.
The effective sample size of the interval [ti-1,ti) is
denoted by ni'.
Let tmi denote the midpoint of [ti-1,ti).
The conditional probability of an event
in [ti-1,ti) is estimated by

and its estimated standard error is

where  .
.The estimate of the survival function at ti is

and its estimated standard error is

The density function at tmi is estimated by

and its estimated standard error is

The estimated hazard function at tmi is

and its estimated standard error is

Let [tj-1,tj) be the interval in which
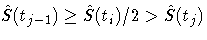 .The median residual lifetime at ti is estimated by
.The median residual lifetime at ti is estimated by

and the corresponding standard error is estimated by

Interval Determination
If you want to determine the intervals exactly,
use the INTERVALS= option in the PROC LIFETEST
statement to specify the interval endpoints.
Use the WIDTH= option to specify the width of the intervals,
thus indirectly determining the number of intervals.
If neither the INTERVALS= option nor the WIDTH= option
is specified in the life table estimation, the number
of intervals is determined by the NINTERVAL= option.
The width of the time intervals is 2, 5, or 10 times
an integer (possibly a negative integer) power of 10.
Let c = log10(maximum event or censored time/number of
intervals), and let b be the largest integer not exceeding c.
Let d=10c-b and let

with I being the indicator function.
The width is then given by
-
width = a ×10b
By default, NINTERVAL=10.
Confidence Limits Added to the Output Data Set
The upper confidence limits (UCL) and the lower confidence
limits (LCL) for the distribution estimates for both the
product-limit and life table methods are computed as

where  is the estimate (either
the survival function, the density, or the hazard
function),
is the estimate (either
the survival function, the density, or the hazard
function),  is the corresponding
estimate of the standard error, and
is the corresponding
estimate of the standard error, and  is the critical value for the normal distribution.
That is,
is the critical value for the normal distribution.
That is,  , where
, where
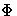 is the cumulative distribution function for the
standard normal distribution.
is the cumulative distribution function for the
standard normal distribution.
The value of  can be specified with the ALPHA= option.
can be specified with the ALPHA= option.
Tests for Equality of Survival Curves across Strata
Log-Rank Test and Wilcoxon Test
The rank statistics used to test homogeneity between the
strata (Kalbfleish and Prentice 1980) have the form of a
c ×1 vector v = (v1,v2, ... ,vc)' with
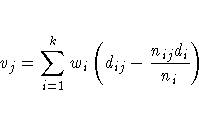
where c is the number of strata, and the estimated
covariance matrix, V = (Vjl), is given by

where i labels the distinct event times, 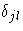 is 1 if
j=l and 0 otherwise, nij is the size of the risk set in the
jth stratum at the ith event time, dij is the number of
events in the jth stratum at the ith time, and
is 1 if
j=l and 0 otherwise, nij is the size of the risk set in the
jth stratum at the ith event time, dij is the number of
events in the jth stratum at the ith time, and

The term vj can be interpreted as a weighted sum
of observed minus expected numbers of failure under
the null hypothesis of identical survival curves.
The weight wi is 1 for the log-rank
test and ni for the Wilcoxon test.
The overall test statistic for homogeneity is
v'V-v, where V-
denotes a generalized inverse of V.
This statistic is treated as having a chi-square distribution
with degrees of freedom equal to the rank of V for
the purposes of computing an approximate probability level.
Likelihood Ratio Test
The likelihood ratio test statistic (Lawless 1982)
for homogeneity assumes that the
data in the various strata are exponentially distributed
and tests that the scale parameters are equal.
The test statistic is computed as

where Nj is the total number of events in the jth
stratum,  , Tj is the total time
on test in the jth stratum, and
, Tj is the total time
on test in the jth stratum, and  .
The approximate probability value is computed by treating
Z as having a chi-square distribution
with c - 1 degrees of freedom.
.
The approximate probability value is computed by treating
Z as having a chi-square distribution
with c - 1 degrees of freedom.
Rank Tests for the Association of Survival Time with Covariates
The rank tests for the association of covariates are
more general cases of the rank tests for homogeneity.
A good discussion of these tests can be
found in Kalbfleisch and Prentice (1980).
In this section, the index  is used to
label all observations,
is used to
label all observations, 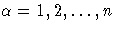 , and the
indices i,j range only over the observations
that correspond to events, i,j = 1,2, ... ,k.
The ordered event times are denoted as t(i),
the corresponding vectors of covariates are denoted as
z(i), and the ordered times, both censored
and event times, are denoted as
, and the
indices i,j range only over the observations
that correspond to events, i,j = 1,2, ... ,k.
The ordered event times are denoted as t(i),
the corresponding vectors of covariates are denoted as
z(i), and the ordered times, both censored
and event times, are denoted as  .
.The rank test statistics have the form

where n is the total number of observations,
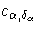 are rank scores,
which can be either log-rank or Wilcoxon rank
scores,
are rank scores,
which can be either log-rank or Wilcoxon rank
scores,  is 1 if the observation
is an event and 0 if the observation is censored,
and
is 1 if the observation
is an event and 0 if the observation is censored,
and  is the vector of covariates
in the TEST statement for the
is the vector of covariates
in the TEST statement for the  th observation.
Notice that the scores,
th observation.
Notice that the scores, 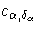 ,
depend on the censoring pattern and that the
summation is over all observations.
,
depend on the censoring pattern and that the
summation is over all observations.
The log-rank scores are

and the Wilcoxon scores are

where nj is the number at risk just prior to t(j).
The estimates used for the covariance
matrix of the log-rank statistics are

where Vi is the corrected sum
of squares and crossproducts matrix for
the risk set at time t(i); that is,

where

The estimate used for the covariance
matrix of the Wilcoxon statistics is
![V = \sum_{i=1}^k
[ a_i (1 - a_i^*)
(2{z}_{(i)}z_{(i)}^' + S_i) -
(a_i^* - a_i)
( a_i x_i{x}_i^' +
\sum_{j=i+1}^k a_j (x_i{x}_j^' +
x_j{x}_i^')
)
]](images/lfteq54.gif)
where

In the case of tied failure times, the statistics
v are averaged over the
possible orderings of the tied failure times.
The covariance matrices are also
averaged over the tied failure times.
Averaging the covariance matrices over the tied orderings
produces functions with appropriate symmetries for the tied
observations; however, the actual variances of the v
statistics would be smaller than the preceding estimates.
Unless the proportion of ties is large,
it is unlikely that this will be a problem.
The univariate tests for each covariate are formed
from each component of v and the corresponding
diagonal element of V as vi2/Vii.
These statistics are treated as coming from a chi-square
distribution for calculation of probability values.
The statistic v'V-v is computed
by sweeping each pivot of the V matrix in
the order of greatest increase to the statistic.
The corresponding sequence of partial statistics is tabulated.
Sequential increments for including a given covariate and the
corresponding probabilities are also included in the same table.
These probabilities are calculated as the tail probabilities
of a chi-square distribution with one degree of freedom.
Because of the selection process, these probabilities
should not be interpreted as p-values.
If desired for data screening purposes, the output
data set requested by the OUTTEST= option can be
treated as a sum of squares and crossproducts matrix
and processed by the REG procedure using the option
METHOD=RSQUARE.
Then the sets of variables of a given size can be found
that give the largest test statistics. Example 37.1
illustrates this process.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.

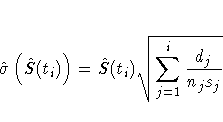

















![]() is the estimate (either
the survival function, the density, or the hazard
function),
is the estimate (either
the survival function, the density, or the hazard
function), ![]() is the corresponding
estimate of the standard error, and
is the corresponding
estimate of the standard error, and ![]() is the critical value for the normal distribution.
That is,
is the critical value for the normal distribution.
That is, ![]() , where
, where
![]() is the cumulative distribution function for the
standard normal distribution.
is the cumulative distribution function for the
standard normal distribution.
![]() can be specified with the ALPHA= option.
can be specified with the ALPHA= option.
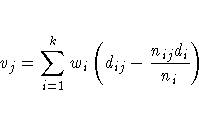









![V = \sum_{i=1}^k
[ a_i (1 - a_i^*)
(2{z}_{(i)}z_{(i)}^' + S_i) -
(a_i^* - a_i)
( a_i x_i{x}_i^' +
\sum_{j=i+1}^k a_j (x_i{x}_j^' +
x_j{x}_i^')
)
]](images/lfteq54.gif)
