Lemurs of Madagascar - Part I
This is the first part of the lemur practical. Write (and draw) your answers on a sheet of paper. Don't forget to write your name and student # on each sheet you submit.
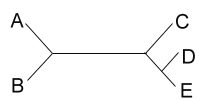
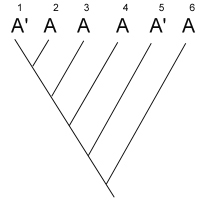
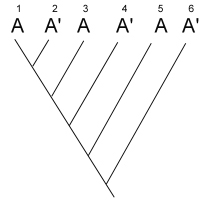
 After studying many characters of these four species, you produce the
unrooted tree shown below:
After studying many characters of these four species, you produce the
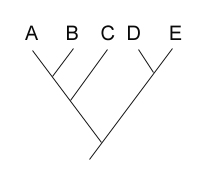
unrooted tree shown below: 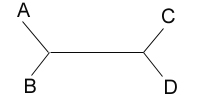 Your next goal is to determine the order of
evolutionary branching. That is, you want to root the tree. You choose an outgroup species
that you think probably has ancestral states of the characters you have studied. However,
due to convergent evolution, your outgroup actually shares many derived characters with
species C. What will your rooted tree look like? How will it differ from the true tree?
Your next goal is to determine the order of
evolutionary branching. That is, you want to root the tree. You choose an outgroup species
that you think probably has ancestral states of the characters you have studied. However,
due to convergent evolution, your outgroup actually shares many derived characters with
species C. What will your rooted tree look like? How will it differ from the true tree? 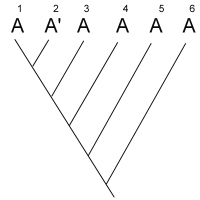
[ 1 2 3 4 5 6 7]
A 1 0 0 0 0 0 1
B 0 1 0 0 0 1 1
C 0 0 1 0 1 1 1
D 0 0 0 1 1 1 1
[ A B C D E]
1 0 1 2 3 3
2 0 2 2 1 3
3 1 3 1 2 3
4 2 3 3 1 2
Optimise the character state distributions onto the tree, using optimisations:
[ 1 2 3 4 5 6]
A.Daubentonia_madagascariensis 0 1 0 0 1 1
B.Oryctolagus_cuninculus 0 1 0 ? 1 0
C.Lemur_catta 1 0 1 1 2 1
D.Indri_indri 1 ? 0 1 2 1
[ Character legend:
1 = number of teeth in tootcomb
0 = toothcomb absent or two teeth
1 = more than two teeth in comb
2 = ever growing incisors
0 = absent
1 = present
3 = ringed tail
0 = absent
1 = present
4 = tonal group alerts
0 = absent
1 = present
5 = skull form
1 = rodentlike
2 = foxlike
6 = opposable thump
0 = absent
1 = present
]
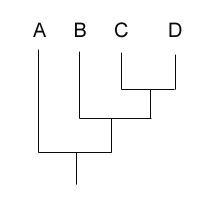
Assume the tree is of the form "(B,(A,(C,D))"
In this final section of part 1, we'll interpret a recent tree of the Lemuridae (Primata). Some background on Madagascar and the lemurs can be
found here.
(Pictures and data by ETI ).
The descriptions of these species are very old (1999) IUCN redbook reports. Note that these
reports follow an old - and taxonomically speaking incorrect - nomenclature for the
taxa.
Download and open a .pdf of our recent lemuridae tree , created in 2010 by Renske Gudde and Jeff Joy.
Download the most recent IUCN taxonomy of lemurs . This is a .csv (comma-separated) file that you can open in Excel
or in a text editor.