Applied Probabilistic Programming. Part I: RStan
Practical Demonstration
Renny Doig
Overview
1. Example Dataset
2. Proposed Statistical Model
3. The Model as a Stan Program
4. Implementation in R
5. Analyzing Results
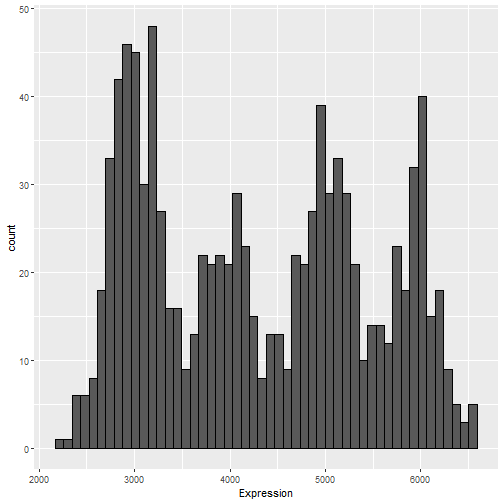
Example Problem Statement
Gene Expression Data
Suppose we have counts for the expression of the MT-ATP6 gene, taken from brain tissues from 1,000 subjects. We would like to make inference about the mean expression level for this gene which accounts for potential difference in means between types of brain tissue. In particular, we assume each sample is taken from one of the four following types:
Cerebellum
Hippocampus
Hypothalamus
Brain stem
Simulated data (modeled on GTEx)
Proposed Statistical Model
Finite Gaussian Mixture Model
A Gaussian mixture model is a linear combination of Gaussian random variables, where each component corresponds to a sub-population and each weight is the probability of belonging to each sub-population
In general, have K components (sub-populations)
A randomly sampled individual belongs to component k∈{1,…,K} with probability αk
Random variable Z used to indicate sub-population: Z∼Categorical(K,αα)
An observation on an individual belonging to component k will be a Y|Z=k∼N(μk,σ)
Finite Gaussian Mixture Model
A Gaussian mixture model is a linear combination of Gaussian random variables, where each component corresponds to a sub-population and each weight is the probability of belonging to each sub-population
In general, have K components (sub-populations)
A randomly sampled individual belongs to component k∈{1,…,K} with probability αk
Random variable Z used to indicate sub-population: Z∼Categorical(K,αα)
An observation on an individual belonging to component k will be a Y|Z=k∼N(μk,σ)
Since Z is discrete it is incompatible with HMC
The workaround is to marginalize out this indicator by evaluating the likelihood as a sum
Y∼K∑k=1αkN(μk,σ)
Bayesian GMM
To make this model Bayesian, we need to assign prior distributions over the component proportions, means, and variance:
Y|αα,μμ,σ∼K∑k=1αkN(μk,σ)αα∼Dirichlet(1)μμ∼Normal(4000,1500)σ2∼InvGamma(3,6002)
Dirichlet is a multivariate extension of the Beta distribution
- Used for a random vectors whose components take values in (0,1) and sum to 1
Bayesian GMM
To make this model Bayesian, we need to assign prior distributions over the component proportions, means, and variance:
Y|αα,μμ,σ∼K∑k=1αkN(μk,σ)αα∼Dirichlet(1)μμ∼Normal(4000,1500)σ2∼InvGamma(3,6002)
Dirichlet is a multivariate extension of the Beta distribution
- Used for a random vectors whose components take values in (0,1) and sum to 1
The resulting posterior distribution is: π(αα,μμ,σ|y)∝n∏i=1[K∑k=1αkN(yi|μk,σ)]N(μμ;4000,1500)IG(σ2;3,6002)D(αα;1)
The Model as a Stan Program
Basic Components of a Stan Program
Our model is specified in a separate .stan file
Written in the Stan probabilistic programming language
Contains up to seven blocks, each serving a different purpose
We only need the three basic blocks:
data{...}: Specify all user inputparameters{...}: Specify any parameters of the model which need to be estimatedmodel{...}: Evaluates the log-posterior
Stan uses this file to automatically construct a Hamiltonian Monte Carlo sampler for this model
Variable Definition
The syntax for defining a variable in Stan is:
type<lower=a><upper=b>[dimension] name;
type: Specifies type of variableBasic types are
intandrealCan also have
vector,matrix, and others
<lower=a>: Specifies the lower bound of the variable; default is unbounded below<upper=b>: Specifies the upper bound of the variable; default is unbounded abovedimension: Specifies dimension of the variableFor
vectorit is the lengthFor n×m
matrixis specified[n,m]
GMM: Data and Parameters
Now we can specify the data and parameters section of our RStan code:
data { int<lower=1> n; // number of observations int<lower=2> K; // number of components vector[n] y; // observations}parameters { simplex[K] alpha; // mixing proportions ordered[K] mu; // means of components real<lower=0> sigma; // standard deviations of components}Identifiability: Without ordering μμ, the model is unidentifiable
E.g. between two iterations, swap μ1 and μ2
- This would leave the likelihood unchanged, and therefore the model is unidentifiable
Imposing an ordering prevents this swapping
Evaluating the Posterior
The log-posterior can be specified in two ways
- Suppose we want to model Y∼N(μ,σ)
Distributional statement:
y ~ normal(mu, sigma);Increment statement:
target += normal_lpdf(y | mu, sigma);targetis a protected keyword and requires special functions to access
These are equivalent statements; can have combination in your
modelblockBoth are vectorized, so if
yis a vector, then it sums over all log-pdf evaluations
Note on Log-Sum-Exponential
Recall in our posterior, we needed to evaluate ∑Kk=1αkN(y|μk,σ)
Since Stan operates on the log-posterior, we need the log of this quantity
In terms of log-densities, this would be written as: log(K∑k=1exp[log(αk)+log(N(y|μk,σ))])
In Stan, if we have a vector
x=[a,b,c], the functionlog_sum_exp(x)will evaluate log(exp(a)+exp(b)+exp(c))This will be used to evaluate the log-posterior
GMM: Model
Recall that the posterior we want to encode is: π(αα,μμ,σ|y)∝n∏i=1[K∑k=1αkN(yi|μk,σ)]N(μμ;4000,1500)IG(σ2;3,6002)D(αα;1)
model { // priors target += gamma_lpdf(1/sigma^2 | 3, 600^2); target += normal_lpdf(mu | 4000, 1500); target += dirichlet_lpdf(alpha | rep_vector(1, K)); // likelihood for(i in 1:n){ vector[K] temp = log(alpha); for(k in 1:K){ temp[k] += normal_lpdf(y[i] | mu[k], sigma); } target += log_sum_exp(temp); }}Implementation in R
The RStan Package
The Stan software is accessed in R through the rstan package
General flow of implementation is
Create .stan file specifying our model
Read data into R and reformat to match the Stan program
Execute the
stanfunctionAutomatically generate a Hamiltonian Monte Carlo sampler
Produce posterior samples, plus diagnostics
Generate plots, numerical summaries, etc.
Note on Compute Canada installation: may require manual installation of V8, Rcpp, or RcppEigen packages
The stan function
The required arguments to the stan function are:
file: A string indicating the path of the .stan file containing the model codedata: A named list with elements matching those specified in the data block of the Stan file
Some optional, but often used arguments:
chains: The number of MCMC chains used (default 4)iter: Total length of each chain (default 2000)init: A list or function used to initialize each chain (default "random")seed: The seed used for random number generation
A full list of parameters can be found in the CRAN documentation
Sampling from the Bayesian GMM
# load the RStan librarylibrary(rstan)# read in dataExpression <- read_csv("../Data/Expression.csv")$Expression# enable parallel computingoptions(mc.cores=5)# set up datan <- length(Expression)K <- 4stan_data <- list(n=n, K=K, y=Expression)Sampling from the Bayesian GMM
# for the rdirichlet functionlibrary(extraDistr)# initial valuesinit_func <- function(){ list(alpha = as.numeric(rdirichlet(1, rep(1, K))), mu = sort(rnorm(K, 4000, 1500)), sigma = 1 / sqrt(rgamma(1, 3, 600^2)))}# run stanfit <- stan(file = "gaussian_mixture.stan", data = stan_data, chains = 5, iter = 1e4, init = init_func, seed = 2)Note: If seed not supplied then Stan will set it randomly, overriding previously set seeds
Analyzing the Results
Examples of Plots
library(bayesplot)mcmc_areas(fit, regex_pars="alpha", prob=0.95) + theme(axis.text = element_text(size=20))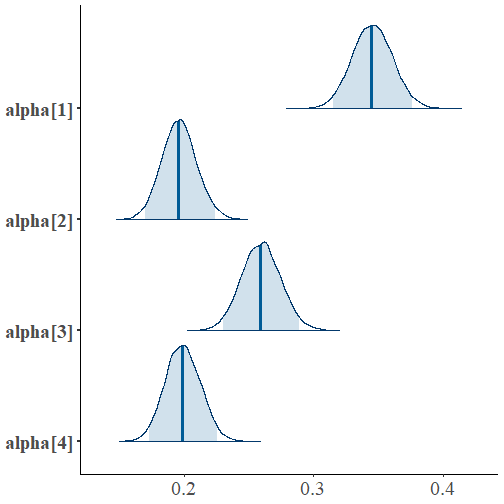
mcmc_areas(fit, regex_pars="mu", prob=0.95) + theme(axis.text = element_text(size=20))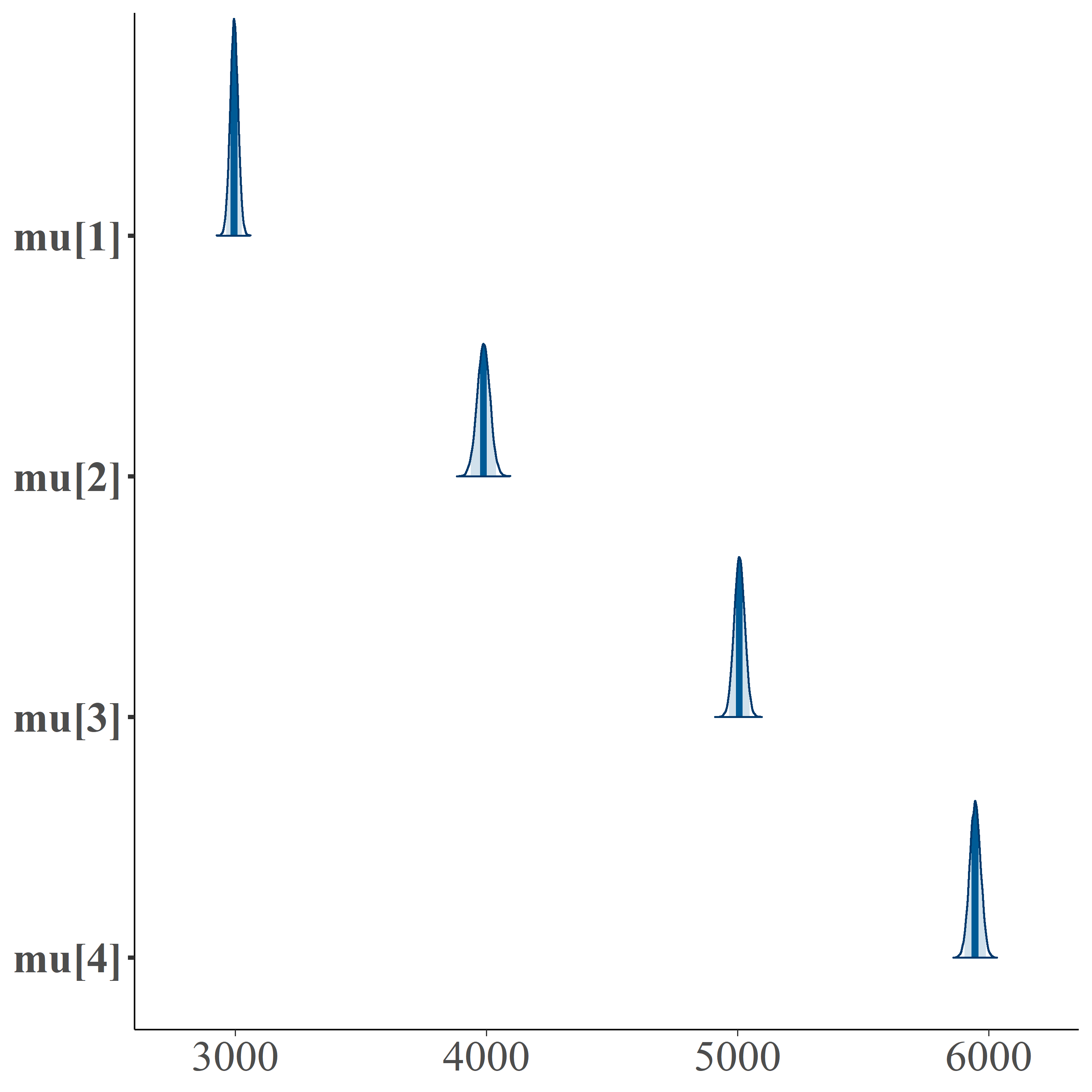
Numerical Summary of Results
RStan also has a summary function for stan-fit objects:
summ <- summary(fit)$summarycolnames(summ) [1] "mean" "se_mean" "sd" "2.5%" "25%" "50%" "75%" [8] "97.5%" "n_eff" "Rhat"round(summ[rownames(summ)!="lp__", c("2.5%", "mean", "97.5%")],2) 2.5% mean 97.5%alpha[1] 0.32 0.35 0.38alpha[2] 0.17 0.20 0.22alpha[3] 0.23 0.26 0.29alpha[4] 0.17 0.20 0.23mu[1] 2963.46 2994.70 3025.78mu[2] 3936.70 3987.60 4038.10mu[3] 4964.72 5006.32 5047.77mu[4] 5902.28 5945.14 5987.92sigma 249.41 263.07 278.16Extracting the Data
It might be preferable to just extract the samples from the stanfit object using the extract function
par_fit <- extract(fit, pars="lp__", include=F)This returns a list of named matrices or vectors of posterior samples
- E.g.
par_fit$alphais a matrix with 4 columns, one for each αk
- E.g.
lp__is the value of the log-posterior at each iterationincludecan be used to invert the meaning ofpars(i.e.lp__is excluded from the extraction)
This can then be converted to a data frame
par_fit <- as.data.frame(par_fit)Concluding Remarks
RStan provides a appealing Bayesian analysis software
Probabilistic programming allows for flexibility; any model that can be written in the Stan languange can be sampled from
A Hamiltonian Monte Carlo sampler is automatically created and executed to sample from the posterior
The package is well-maintained; no debugging of the algorithm itself necessary
These slides can be found here: https://www.sfu.ca/~rennyd/RStanTutorial/tutorial.html
- Supporting files can be found in the parent directory